Population-based epidemiologic studies are powerful tools linking clinical, genetic, and immunologic data to identify potential novel mechanisms of disease.
Genomewide Association Study (GWAS) of NLRP3 Inflammasome-Induced Cytokines in Virally Suppressed People with HIV (PWH)
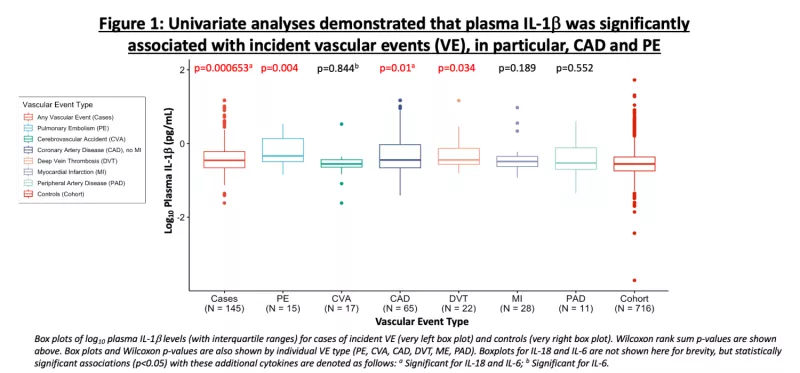
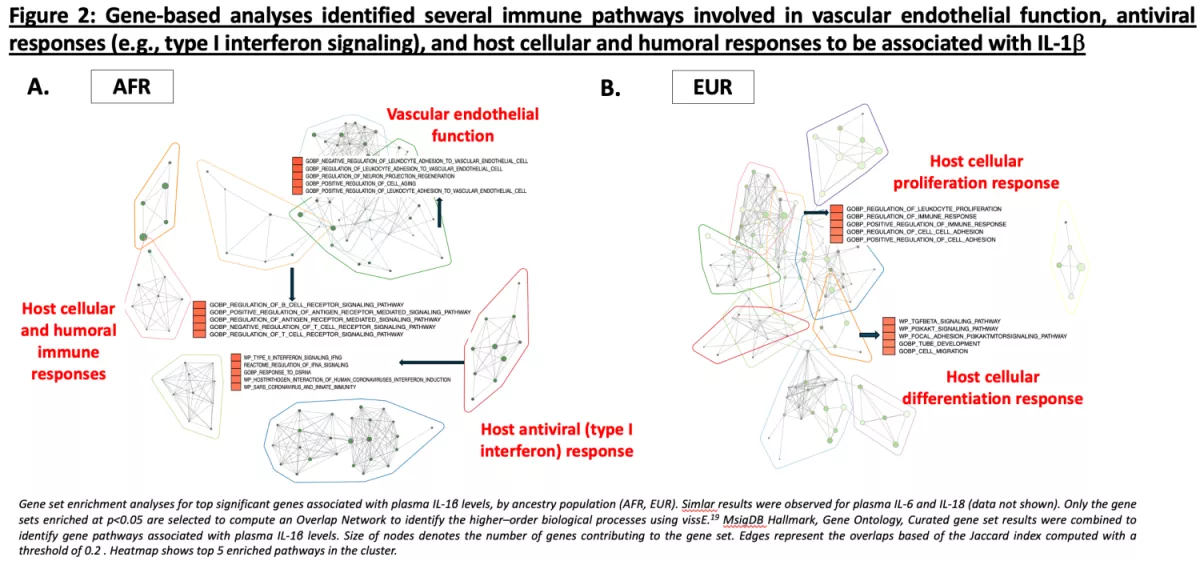
Despite effective antiretroviral therapy (ART), PWH experience higher rates of morbidity and mortality compared to age-matched people without HIV (PWoH). Chronic inflammation due to persistent virus (“the HIV reservoir”) may drive these outcomes. Indeed, markers of inflammation are significantly higher in PWH compared to PWoH and predict morbidity and mortality even during ART. Plasma interleukin (IL)-6 levels are amongst the strongest predictors of mortality in U.S.-based HIV cohorts. Recent data suggests that IL-1β, upstream of IL-6, may be the major driver of this risk. IL-6, IL-1β, and IL-18 are cytokines induced by the NOD-like receptor 3 (NLRP3) inflammasome, which plays a critical role in the innate immune response to pathogens. Leveraging samples from the U.S. Military HIV Natural History Study, we conducted a case-cohort study to (1) determine the association between circulating plasma NLR3 inflammasome-induced cytokines in relation to incident vascular events (e.g., heart disease, stroke, etc.) and (2) to identify potential host genetic variants associated with these levels.
GWAS of Long COVID
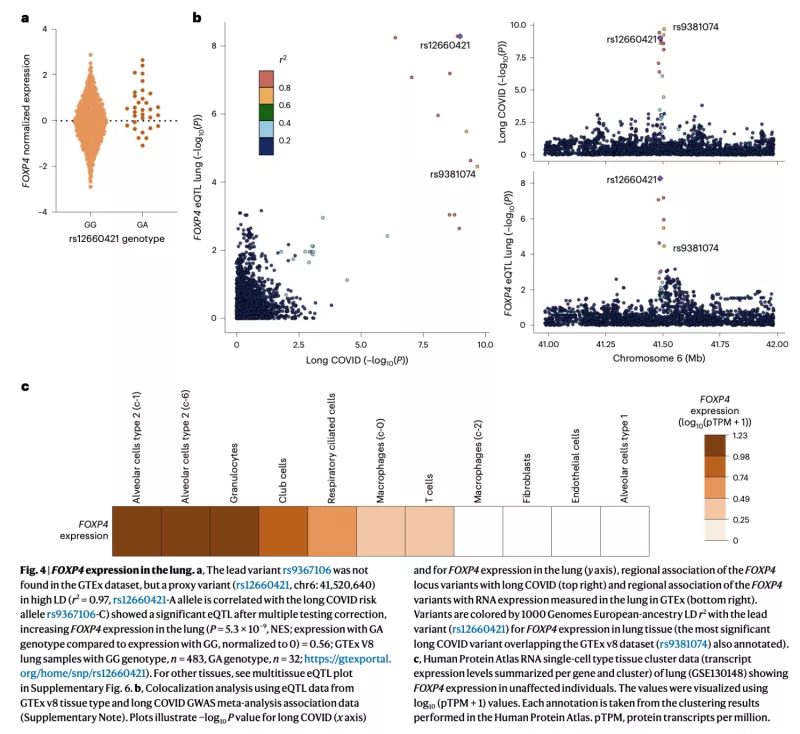
Infections can lead to persistent symptoms and diseases such as shingles after varicella zoster or rheumatic fever after streptococcal infections. Similarly, severe acute respiratory syndrome coronavirus 2 (SARS‑CoV‑2) infection can result in long coronavirus disease (COVID), typically manifesting as fatigue, pulmonary symptoms and cognitive dysfunction. The biological mechanisms behind long COVID remain unclear. In collaboration with the international COVID-19 Host Genetics Initiative, we performed a genome-wide association study for long COVID including up to 6,450 long COVID cases and 1,093,995 population controls from 24 studies across 16 countries. We discovered an association of FOXP4 with long COVID, independent of its previously identified association with severe COVID-19. The signal was replicated in 9,500 long COVID cases and 798,835 population controls. Given the transcription factor FOXP4’s role in lung physiology and pathology, our findings highlight the importance of lung function in the pathophysiology of long COVID.
GWAS of Biomarkers Associated with Mortality in PWH on ART in Uganda
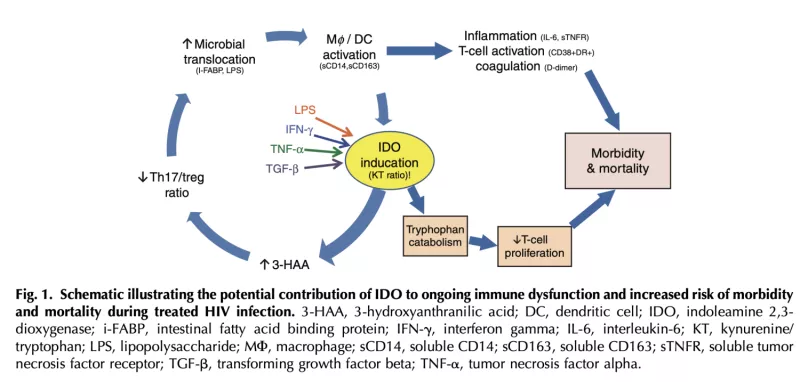
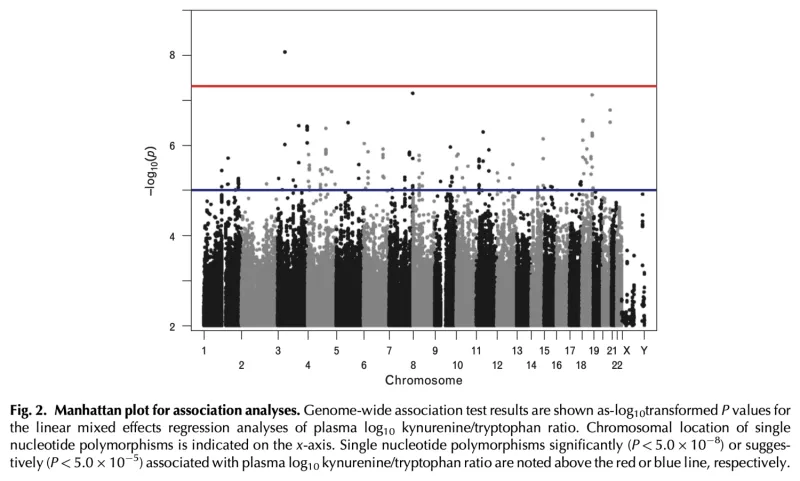
Plasma kynurenine/tryptophan (KT) ratio, a biomarker of indoleamine 2,3-dioxygenase-1 (IDO) activity, is a strong independent predictor of mortality in HIV-infected Ugandans initiating antiretroviral therapy (ART) and may play a key role in HIV pathogenesis. We performed genotyping to identify variants associated with plasma KT ratio during 6-12 months of suppressive ART in Ugandans from the Uganda AIDS Rural Treatment Outcomes (UARTO) and Antiretrovirals in Kaposi Sarcoma (ARKS) cohorts. We evaluated >16 million single nucleotide polymorphisms (SNPs) in association with log10KT ratio using linear mixed models adjusted for cohort, gender, pregnancy, and ancestry. Among 597 Ugandans, we identified several SNPs in TNF, IFNGR1, and TLR4 that were associated with log10KT ratio (P<5.0E5). An intergenic polymorphism between CSPG5 and ELP6 was genome-wide significant, while several others exhibited suggestive associations (P<5.0E7), including genes encoding protein tyrosine phosphatases (PTPRM, PTPRN2) and the vitamin D metabolism gene, CYP24A1. Several of these were associated with markers of inflammation, coagulation, and monocyte activation. These findings highlight a potentially important role of IFN-γ, TNF-α, and toll-like receptor signaling in determining IDO activity and subsequent mortality risk in HIV-infected ART-suppressed Ugandans. These results also identify potential novel pathways involved in IDO immunoregulation.
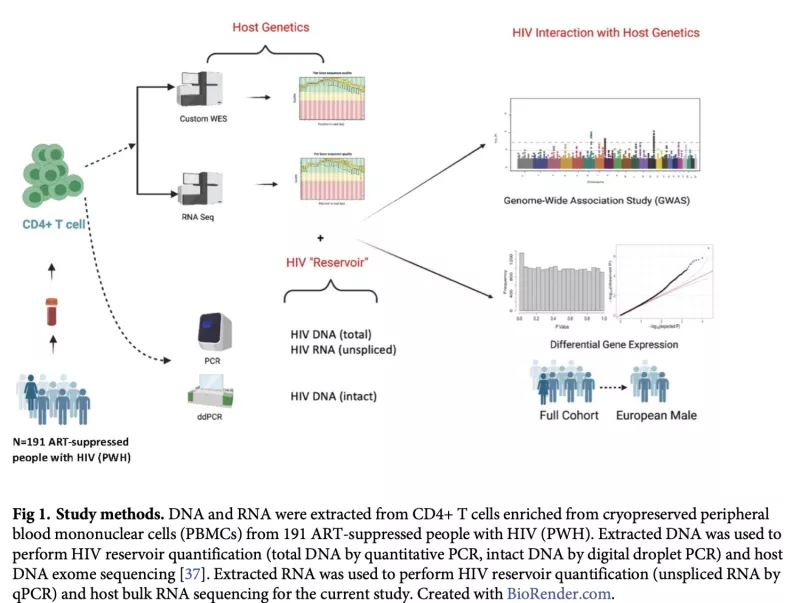
Whole Exome Sequencing (WES) Analysis to Identify Genetic Variants Associated with HIV Reservoir in PWH on ART
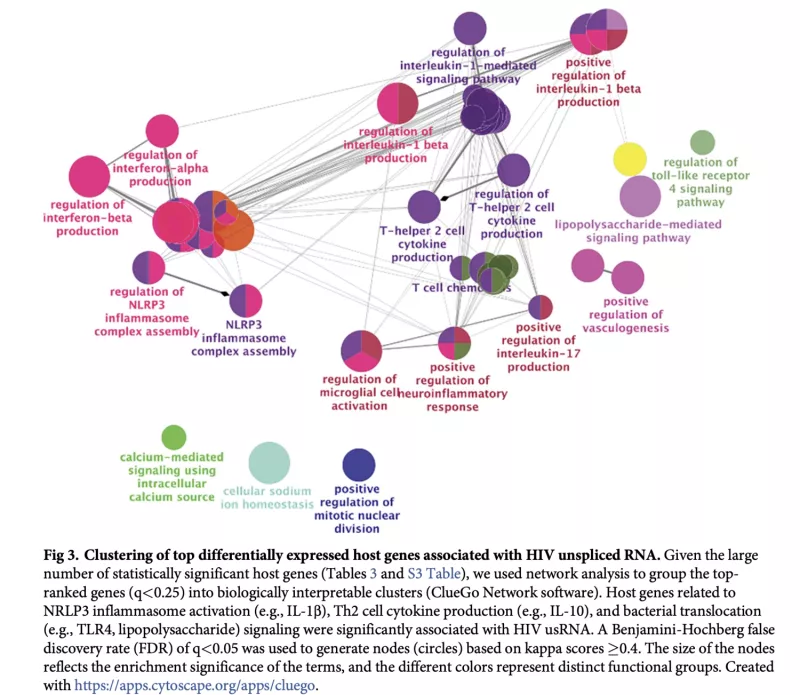
The major barrier to an HIV cure is the HIV reservoir: latently-infected cells that persist despite effective antiretroviral therapy (ART). There have been few cohort-based studies evaluating host genomic or transcriptomic predictors of the HIV reservoir. We performed host RNA sequencing and HIV reservoir quantification (total DNA [tDNA], unspliced RNA [usRNA], intact DNA) from peripheral CD4+ T cells from 191 ART-suppressed people with HIV (PWH) from the UCSF SCOPE HIV cohort. We identified 17 host genes for which lower expression was associated with higher residual transcription (HIV usRNA). These included novel associations with membrane channel (KCNJ2, GJB2), inflammasome (IL1A, CSF3, TNFAIP5, TNFAIP6, TNFAIP9, CXCL3, CXCL10), and innate immunity (TLR7) genes (FDR-adjusted q<0.05). Gene set enrichment analyses further identified significant associations of HIV usRNA with TLR4/microbial translocation (q=0.006), IL-1/NRLP3 inflammasome (q=0.008), and IL-10 (q=0.037) signaling. Protein validation assays using ELISA and multiplex cytokine assays supported these observed inverse host gene correlations, with P3H3, IL-10, and TNF-a protein associations achieving statistical significance (p<0.05). Our analyses also identified known tumor suppressor genes, P3H3 and NBL1, that were significantly associated with smaller total DNA viral reservoir size. These data suggest that host gene expression may vary in response to the transcriptionally active reservoir and that changes in cellular proliferation genes may influence the size of the HIV reservoir. These findings add important data to the limited host genetic HIV reservoir studies to date.